What's new in USEARCH v11
See also
What's new in version 10
New features
Machine learning
Improved cross-talk detection
Novel alpha diversity metrics
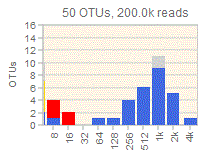
Octave plots for visualizing alpha diversity
Statistical significance of diversity differences between groups
Cross-validation by identity (CVI)
New commands
calc_lcr_probs Calculate lowest common rank probabilities
cluster_tree Construct clusters from tree using distance cutoff
distmx_split_identity Split distance matrix into test/training pair for CVI
fastx_syncpairs Sort forward and reverse reads into the same order
fastx_trim_primer Remove primer-binding sequence from FASTx file
forest_classify Classify data using random forest
forest_train Train random forest classifier
nbc_tax Predict taxonomy using RDP Naive Bayesian Classifier algorithm
otutab_binary Convert OTU table with counts to presence(1) / absence(0)
otutab_forest_classify Classify samples using random forest
otutab_core Identify core microbiome in OTU table
otutab_forest_train Train random forest classifier on OTU table
otutab_otus Extract OTU labels from OTU table
otutab_rare Sub-sample OTU table to same number of reads per sample
otutab_samples Extract sample labels from OTU table
otutab_select Identify OTUs which are informative (correlate with metadata)
otutab_xtalk Identify cross-talk using improved algorithm ( UNCROSS2 )
search_pcr2 In-silico PCR, search for matches to primer pair
subtree Extracts subtree under given node
tabbed2otutab Convert read mapping file (read+OTU) to OTU table
tree_subset Extract subset of tree for given set of leaf labels

