Chimeras
See also
Quality control for OTU sequences
Checking for chimeras in OTU sequences
Fake chimeras
Low-divergence chimeras are common
Chimeras are sequences formed from two or more biological sequences joined together. Amplicons with chimeric sequences can form during PCR . Chimeras are rare with shotgun sequencing, but are common in amplicon sequencing when closely related sequences are amplified. Although chimeras can be formed by a number of mechanisms, the majority of chimeras are believed to arise from incomplete extension. During subsequent cycles of PCR, a partially extended strand can bind to a template derived from a different but similar sequence. This then acts as a primer that is extended to form a chimeric sequence ( [Smith et al. 2010] , [Thompson et al., 2002] , [Meyerhans et al., 1990] , [Judo et al., 1998] , [Odelberg, 1995] ).
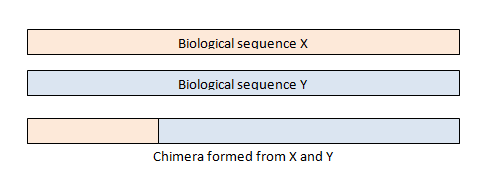
A chimeric template is created during one round, then amplified by subsequent rounds to produce chimeric amplicons. In 16S sequencing, we typically find that only a small fraction of reads is chimeric, perhaps of the order of 1% to 5%. However, when reads are clustered into groups of unique sequences or into OTUs, then we often find that a much larger fraction is chimeric (see Tolstoy's paradox ). This is a challenging problem in sequence analysis because chimeras often have low divergence , i.e. are very similar to one of their parents, so are difficult to distinguish from true biological sequences.
It turns out that it is impossible in principle to distinguish chimeras from correct sequences, even when there are no sequence errors and the reference database is complete. This is a very surprising, almost shocking, result which is reported in the UCHIME2 paper . The reason is "f ake models ", where a correct sequence can be constructed as a chimera from two other correct sequences. Chimeras can have identical sequences to valid genes, so it is impossible for an algorithm to distinguish the two cases from a sequence alone. Fake models are common in practice, hence the problem.
Reference (please cite)
R.C. Edgar (2016), UCHIME2: improved chimera prediction for amplicon sequencing, https://doi.org/10.1101/074252
• UCHIME2 algorithm, improved chimera detection
• "Fake" chimeras are common, valid biological sequences matching two-parent model
• Perfect chimera filtering impossible even with complete and correct reference
• Realistic chimera benchmark

