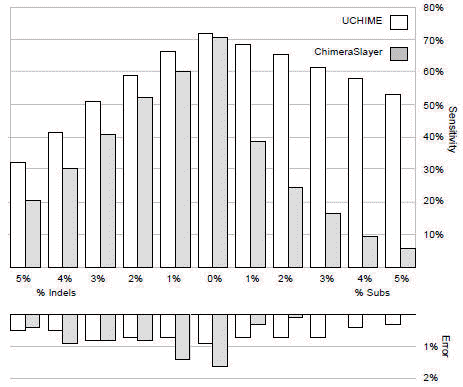
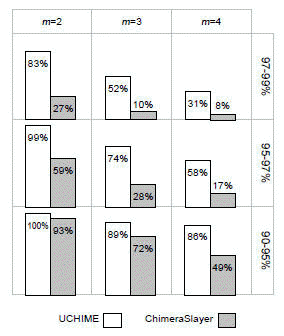
UCHIME vs. ChimeraSlayer
See also
Chimera benchmark home
UCHIME and ChimeraSlayer
Below are Figs. 5 (left) and Fig. 4 (right) from Edgar et al. (2011) . Fig 5. shows sensitivity and error rate measured on a set of simulated chimeric sequences of length 200 with from 0% to 5% errors introduced. The simulated chimeras have two segments. Fig 4. shows sensitivity on a different set of simulated chimeras with two , three or four segments (columns). Rows are divergence ranges (divergence is the identity of the chimera with the most similar parent sequence). These results show that UCHIME (reference database mode) has significantly better sensitivity and lower error rate than ChimeraSlayer.