sintax_summary command
 See also
See also
sintax command
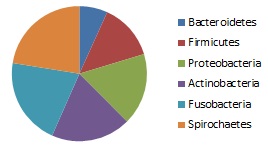
Creates a summary report for sintax predictions giving the frequencies of each named taxon at a given rank. For example, at phylum rank, the report might show 25% Bacteroidetes, 18% Firmicutes etc.
Input is an tabbed text output file from the sintax command (tabbedout option).
The rank option is required, it is a single letter specifying the rank, e.g. p for phylum. See taxonomy annotations for value letters.
An OTU table can be specified using the otutabin option. This will generate a report with frequencies for each sample and over all samples combined. The table must be in QIIME classic format . OTU identifiers in the OTU table must exactly match the labels in the sintax output file.
If size annotations are found in the sequence labels in the sintax output file, they are taken into account in calculating frequencies.
The output from sintax_summary can be imported into a spreadsheet or charting program for making figures.
Example
usearch -sintax_summary sintax.txt -otutabin otutab.txt \
-output phylum_summary.txt -rank p

