Connected components of a graph
See also
cluster_edges command
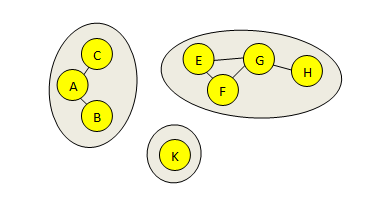
A graph is a set of points (called "nodes") in which pairs of nodes are connected by lines (called "edges"). In the figure below, there is a graph with eight nodes, six edges and three connected components.
A connected component is a maximal set of nodes such that a path can be found between any two nodes. Connected components form disconnected "islands" in a graph because by definition there can be no edge between two components.
Finding the connected components of a graph is equivalent to agglomerative clustering with single linkage .
In USEARCH, finding connected components is supported by cluster_agg (input is sequences; use - linkage min), cluster_aggd (input is a distance matrix ; use -linkage min) and by cluster_edges (input is a tabbed text file specifying the edges).