UPARSE-OTU algorithm
See also
Defining and interpreting OTUs
UPARSE-REF algorithm
UPARSE home page
OTU benchmark results
UPARSE pipeline
cluster_otus command
Should I use UPARSE or UNOISE?
Introduction
The UPARSE-OTU algorithm constructs a set of OTU representative sequences from NGS amplicon reads. It is implemented in the cluster_otus command. Reads should be pre-processed to overlap paired reads (if appropriate), strip barcodes, perform quality filtering and global trimming . Post-processing is needed to map reads to OTUs and construct a OTU table. See UPARSE pipeline for detailed discussion of practical issues. This page describes the OTU clustering algorithm itself. Assigning reads to OTUs is a separate task which is not addressed by UPARSE-OTU. See otutab command and defining OTUs for more details.
Input sequences
Input to UPARSE-OTU is a set of sequences. Each sequence is marked with an integer value indicating its abundance. In practice, the abundance is usually the number of reads having a given unique sequence, but it could also be the predicted abundance of an amplicon after a denoising step.
Clustering criteria
The goal of UPARSE-OTU is to identify a set of OTU representative sequences (a subset of the input sequences) satisfying the following criteria.
1. All pairs of OTU sequences should have <97% pair-wise sequence identity.
2. An OTU sequence should be the most abundant within a 97% neighborhood.
3 . Chimeric sequences should be discarded.
4. All non-chimeric input sequences should match at least one OTU with >= 97% identity. 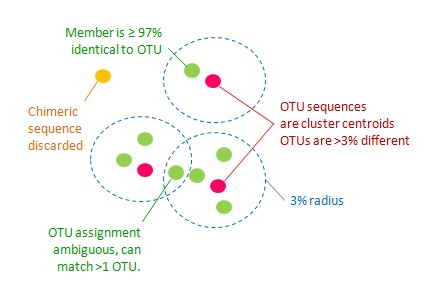
Greedy clustering
UPARSE-OTU uses a greedy algorithm to find a biologically relevant solution, as follows. Since high-abundance reads are more likely to be correct amplicon sequences, and hence are more likely to be true biological sequences, UPARSE-OTU considers input sequences in order of decreasing abundance. This means that OTU centroids tend to be selected from the more abundant reads, and hence are more likely to be correct biological sequences.
Each input sequence is compared to the current OTU database, and an maximum parsimony model of the sequence is found using UPARSE-REF (figure below). There are three cases. (a) The UPARSE-REF model is >= 97% identical to an existing OTU, (b) the model is chimeric, or (c) the model is <97% identical to any existing OTU. In case (a), the input sequence becomes a member of the OTU. In case (b), the input sequence is discarded. In case (c), the input sequence is added to the database and becomes the representative sequence (centroid) of a new OTU.
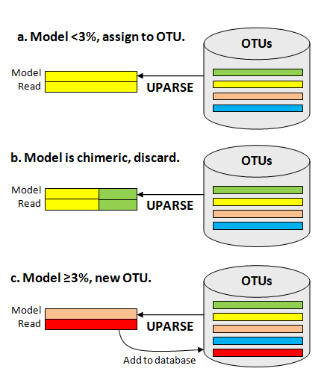
Reference
Edgar, R.C. (2013) UPARSE: Highly accurate OTU sequences from microbial amplicon reads, Nature Methods [ Pubmed:23955772 , dx.doi.org/10.1038/nmeth.2604 ].

