See also
Taxonomy benchmark home
Validating taxonomy classifiers
Training taxonomy classifiers
UTAX algorithm
Splitting a taxonomy reference set
Defining "accuracy" of a taxonomy classifier
Taxonomy classification errors
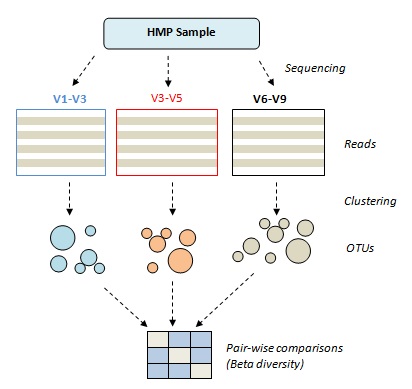
Can we make OTUs from different V regions?
This test uses samples from the Human
Microbiome Project that were sequenced using three different primer sets
targeting different V regions (V1-V3, V3-V5 and V6-V9). See
HMP sample test results.
For a given sample, OTUs are constructed for each set of reads and pair-wise comparisons are made using weighted Jaccard similarity. If the clustering method works well, the similarity will be high, indicating that the OTUs made from each region are similar. Ideally, the similarity would be 100% because the original sample was identical for each set of reads, but even if the OTU construction is ideal we expect <100% similarity in practice because of sampling effects (rare taxa may not appear in a given set of reads).