See also
Taxonomy benchmark home
Validating taxonomy classifiers
Training taxonomy classifiers
UTAX algorithm
Splitting a taxonomy reference set
Defining "accuracy" of a taxonomy classifier
Taxonomy classification errors
Can we make OTUs from different V regions?
This test uses samples from the Human
Microbiome Project that were sequenced using three different primer sets
targeting different V regions (V1-V3, V3-V5 and V6-V9). See
HMP sample validation for details.
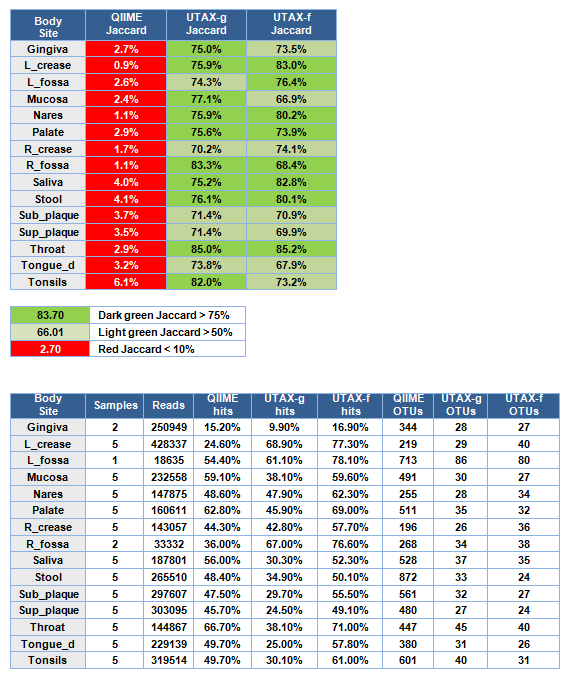
Results are shown in the tables below. The top table shows the weighted Jaccard similarity averaged over samples from each body site. In all cases, the similarity of QIIME closed-ref OTUs are < 10% while the similarity of UTAX OTUs at family and genus level are 67% to 85%.
In the lower table, "hits" indicates the fraction of reads that were classified into OTUs.