See also
Taxonomy benchmark home
Validating taxonomy classifiers
Training taxonomy classifiers
UTAX algorithm
Splitting a taxonomy reference set
Defining "accuracy" of a taxonomy classifier
Taxonomy classification errors
Can we make OTUs from different V regions?
This test uses full-length 16S sequences from the
Greengenes database.
Three different segments are extracted: V3, V4 and V3-V5 using primer pairs
commonly used in tag sequencing experiments. See
Greengenes fragment test for details.
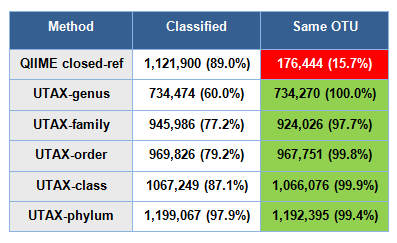
Results are shown in the table below.
Classified is the number of genes where all three fragments (V4, V5 and V3-V5) were classified. In the case of QIIME closed-ref, it is notable that more than 10% are not classified, considering that the reference database is Greengenes. This happens because the reference database is Greengenes clustered at 97% identity rather than the full Greengenes set.
Same OTU is the number of of genes where all three fragments (V3, V4 and V3-V5) were assigned to the same OTU.
This test should favor the QIIME closed-reference method because the Greengenes database is used as both the query and reference set, while UTAX uses the RDP training set as a reference. However, QIIME only identifies 15.7% of fragments as belonging to the same gene and thus fails in most cases (84.3%). This is consistent with the very poor results for QIIME closed-ref on the HMP sample test where the similarity between identical samples sequenced by different V-regions ranges from 6.1% to 0.9%.