
NR protein search: metagenomics benchmark
Methods
The NCBI non-redundant protein database (NR) was downloaded
Dec 2013. This is a 15 Gb FASTA file containing 38.5 M proteins.
The query set was 1,000 454 metagenomic shotgun reads, chosen at random from SRA run SRR606808. Average read length is 421nt.
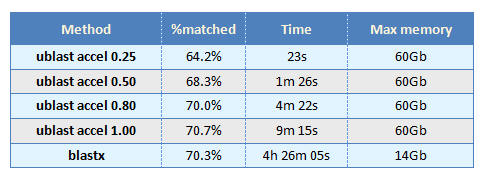
The -dbaccelpct 80 option was used to reduce the size of the USEARCH udb database, which otherwise would have exceeded the available RAM (64 Gb).
A read was considered to be matched to the database if it had at least one hit with E-value of 1e-3 or less. The E-values calculated by ublast and blastx can differ by several orders of magnitude but this effect does not significantly bias the results in favor of either program.
The udb database load time was excluded from the timing, on the assumption that in practice, the load time is small compared to the search time so this gives a more realistic assessment of the relative search speeds.
See also: hardware configuration.
Command lines
usearch -ublast reads.fa -db nr.udb -accel 0.8 -evalue 1e-3 -threads 6
-blast6out hits.b6
blastx -query reads.fa -db nr -evalue 1e-3 -outfmt 6 >
hits.b6