
Greengenes clustering benchmark
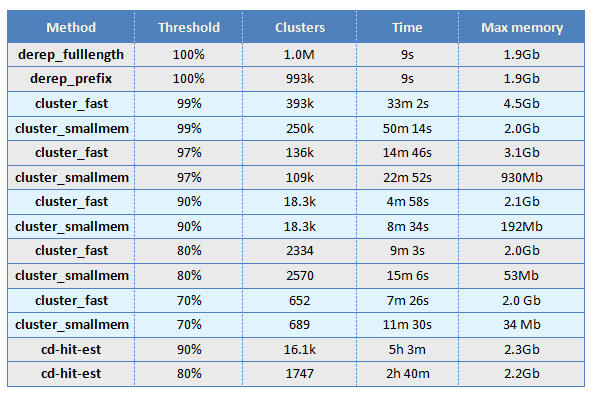
Methods
Input was the
Greengenes
16S RNA sequence database, version 13.5. This is a
1.7Gb FASTA
file, containing 1,262,986 (1.3M) sequences.
USEARCH clustering was tested using the derep_fulllength, derep_prefix, cluster_fast and cluster_smallmem commands in USEARCH v7.0.1090, 64-bit build for Linux.
CD-HIT clustering was tested using cd-hit-est v4.6. At 100% identity, the cd-hit run had not finished clustering after several days and was canceled; the full results are therefore not known.
See also: hardware configuration.
Command lines
usearch -derep_fulllength gg.fa -threads 6 -output full.fa
usearch -derep_prefix gg.fa -threads 6 -output prefix.fa
usearch -cluster_fast gg.fa -id 0.97 -threads 6 -centroids fast97.fa
usearch -cluster_smallmem gg.fa -id 0.97 -centroids small97.fa
cd-hit-est -i gg.fa -n 5 -M 0 -T 6 -c 0.8 -o gg80