See also
fastq_mergepairs command
fastq_mergepairs options
Validating merged reads to check for problems
Using the tabbedout file to investigate
merging problems
Trouble-shooting problems with fastq_mergepairs
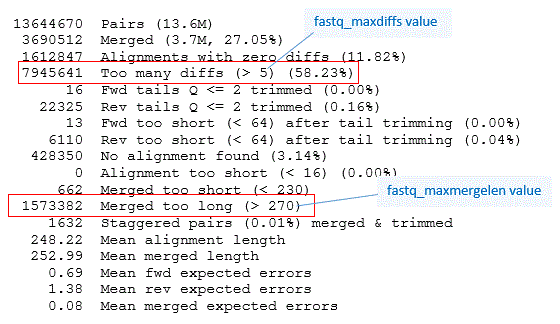
Below is an example report produced by the -report option of fastq_mergepairs. This information is also shown on the terminal (standard error output stream). The options -fastq_minmergelen 230 -fastq_maxmergelen 270 were used because these are 2 x 250 reads of amplicons generated by three different primer pairs including V4, V3-V4 and V4-V5. Using the length range 230 to 270 selects the V4 reads.
For each parameter used in pre-processing, alignment, merging and filtering the report shows how many pairs were successfully processed or discarded. The parameter value is also shown. For example, 7.9M reads (58%) were discarded because the alignment had >5 mismatches (parameter set by the fastq_maxdiffs option).
Here we have long overlaps, shown by the mean
alignment length of 248. Mis-alignments are therefore very unlikely, and it
would be reasonable to increase the -fastq_maxdiffs and -fastq_maxdiffspct
values to increase the number of merged pairs. Quality filtering will take
care of discarding reads where many mismatches induce a large number of
expected errors. This doesn't necessarily happen -- e.g., if low quality
base calls in R2 are mismatches against high-quality base calls in R1 then
the merged Q scores can still be high.
If you think that too many
reads are being discarded, then you can
use the tabbedout file to investigate further.