| USEARCH manual > dereplication > substring dereplication using version 5.2 | |||||||||||||||
| substring dereplication using version 5.2 | |||||||||||||||
| As noted here, USEARCH v6.0 does not support substring dereplication. However, this feature was implemented in v5.2, which is still available and will be supported for the foreseeable future. A 32-bit binary for v5.2 can be obtained from the download page by using the drop-down list:
Sequences should be presorted by decreasing length, this can be done using sortbylength in v6 or the equivalent -sort command in v5. Here is an example command-line. usearch5.2.32_i86linux32 -cluster seqs.fasta -derep_subseq
-minlen 64 \ Supported output options include -seedsout (FASTA file for the unique sequences, equivalent to -centroids in v6) and -uc (same as the -uc option in v6). Following are options that should usually be set. See indexing options for an explanation.
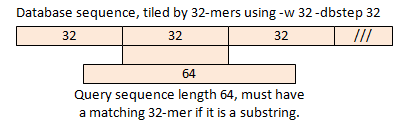
How to choose parameters Suppose we tile the database with non-overlapping 32-mers using -w 32 and -dbstep 32. Then a substring of length 64 or more must have at least one matching 32-mer, as shown in the figure below.
As this example shows, if the minimum sequence length is L, we use a word length w <= L/2 and -dbstep w, then we have (1) minimized memory use (we use the fewest possible words by tiling the database) and (2) a substring is guaranteed to have at least one word in common. Note that all words in the query are considered; the -dbstep option only applies to the database sequences. If w is also large enough that random word matches are very unlikely, then this algorithm is usually very effective, but is still heuristic strictly speaking unless we use -maxrejects 0. There is one small problem I
glossed over. If -dbstep 32 is used, the database sequence is not fully
covered unless its length is a multiple of 32. There is usually a fragment
at the end of length < 32, indicated by /// in the figure. There may be some
false negative matches for substrings that match such fragments. This can be
mitigated by using a dbstep value that is smaller than w, say w/2. |