
Greengenes: High-identity nucleotide search benchmark
Description
The Greengenes search benchmark tests high-identity nucleotide search. This test should be considered informative
rather than rigorous, because rigorous
benchmarking of USEARCH against BLAST is not possible.
The query set was one set of 16S amplicon reads chosen at random from the Human Microbiome Project, SRA run SRR045567. After filtering, the query set contained 5,893 reads with an average length of 503nt.
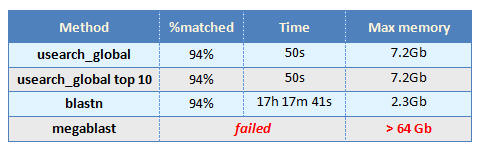
The %matched column shows the fraction of reads with a hit having at least 90% identity. In the case of blastn, the hit must cover at least 90% of the query sequence.
The "top 10" variant of usearch uses the termination options -maxaccepts 10 -maxrejects 100 to simulate a situation where several top hits are desired rather than a single top hit. In this case, increasing the number of hits per query had no measurable effect on the execution time or memory use.
Command lines
usearch_global reads.fa -db gg.udb -id 0.9 -strand plus -threads 6 -blast6out
hits.b6
usearch_global reads.fa -db gg.udb -id 0.9 -strand plus -threads 6 -blast6out
hits.b6 \
-maxaccepts 10 -maxrejects 100
blastn -query reads.fa -db gg -num_threads 6 -outfmt 6 > hits.b6
blastn -task megablast -use_index true -index_name ggmb -query reads.fa \
-db gg -num_threads 6 -outfmt 6 > hits.b6