How to make high-quality protein structure alignment figures
First, use the alignpair command to rotate the query structure so that it is aligned with the target structures, e.g.:
reseek -alignpair 1hhs.pdb -input2 7c2k.pdb -aln 1hhs_7c2k.txt \
-output 1hhs_rotated.pdb
Load 72ck_rotated.pdb into Pymol using File, Open .
Load 1hhs.pdb by using File, Open a second time in the same window.
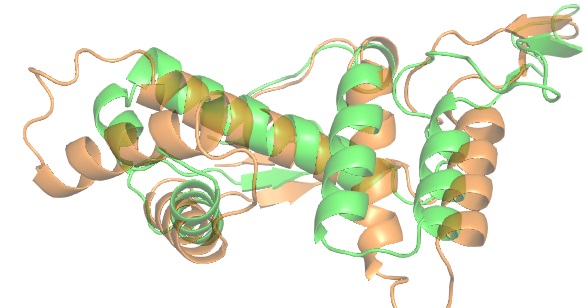
You should see something like this:
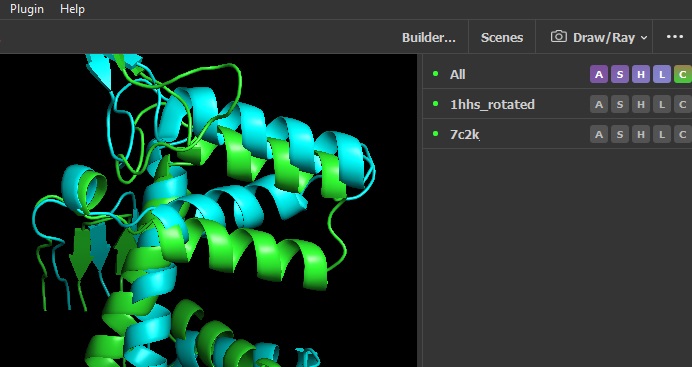
For best image quality, select Display, Quality > Maximum Quality .
Set the background color using Display, Background , probably White or Alpha Checker is best. The Alpha Checker option gives you a transparent background which is helpful if you're going to be adding annotations in image editing software such as Gimp or Inkscape .
Choose colors for the structures by clicking the [C] buttons.
Some transparency may make the superposition clearer (...literally). For example, if you have chosen a cartoon representation, then you can go to Setting, Transparency > Cartoon , something like 40% transparency looks pretty good.
Now you should should have something like this, which you can save using choose File, Export Image As .