Score options
Command-line options for the UCHIME scoring function are shown in the
following table; see below for explanation.
| Option |
|
Default |
Description |
|
‑minh |
|
0.28 |
Minimum score (h) to be classified as chimera. Increasing this value tends to increase the
number of false positives (and also sensitivity). |
|
‑xn |
|
8.0 |
Weight of "no" vote (β). Increasing this value tends
to decrease the number of false positives (and also sensitivity). Must be
> 1. |
|
‑dn |
|
1.4 |
Pseudo-count prior for "no" votes. (n). Increasing this
value tends to decrease the number of false positives (and also
sensitivity). Must be > 0. |
|
‑mindiffs |
|
3 |
Minimum number of diffs in a segment. Increasing this
value tends to reduce the number of false positives while reducing
sensitivity to very low-divergence chimeras. Must be >
0. |
|
‑mindiv |
|
0.8 |
Minimum divergence, i.e. 100% - identity between the query
and closest reference database sequence. Expressed as a percentage,
so the default is 0.8%, which allows chimeras that are up to 99.2%
similar to a reference sequence. This value is chosen to improve
sensitivity to very low-divergence chimeras as needed to achieve a
high score on the SIM2 benchmark (see UCHIME paper). I
generally recommend increasing this value to, say, 1.5 to further
reduce the number of false positives, which allows reducing the
‑minh option to improve sensitivity to higher-divergence chimeras.
Must be > 0. |
UCHIME alignment
A typical
UCHIME alignment. The query sequence is Q, the putative parent
sequences are A and B. The true parent sequences may not be present
in the reference database, in which case a closely related sequence
("step-parent") might be used instead.
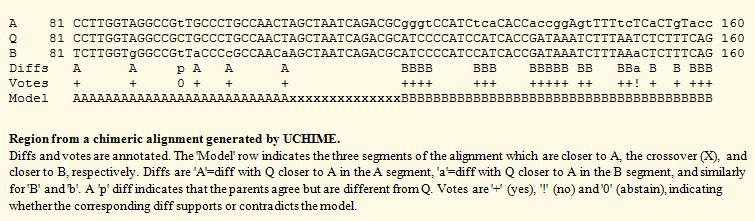
Diffs and votes
In a typical
alignment, most columns are identities q=a=b, where q, a and b are
letters from Q, A and B respectively. A column in which at least
one sequence differs from the other two is called a diff. Diffs can
be considered as votes for or against the model. For example, a
diff q=a, q≠b increases the distance d(Q,B) while leaving d(Q,A)
unchanged. If such a diff is found in the segment that is closer to
A, it can be regarded as a "yes" vote supporting the model; if it
is found in the segment that is closer to B then it contradicts the
model and is regarded as a "no" vote. A diff in which all three
sequences differ or in which a=b, q≠a, q≠b increases the
distance of Q to both A and B and is regarded as an "abstain" vote
that neither supports nor contradicts the model. Let Yg, Ng and Ag
be the total number of yes, no and abstain votes in segment g of
the model, where g is L (left) or R (right). If YL >
NL > NL and YR >
NR, the alignment is chimeric and the model is closer to
Q than A or B alone. The number of diffs may be very small in more
challenging cases. For example, in a 16S experiment using 200nt
reads, clusters of radius ~3% might be used in an attempt to
identify species. It would then be important to identify chimeras
with divergences as low as ~2%, which could have a few as four
diffs with their closest parents. In such cases, the small amount
of evidence available should increase the uncertainty of the
classification. The ‑mindiffs option sets the minimum number of
diffs that must be present in a segment; increasing this value
tends to reduce the number of false positives while reducing
sensitivity to very low-divergence chimeras.
UCHIME scoring function
Each segment g (left and right
of the cross-over) is assigned a score:
Hg = Yg /
(β (Ng + n) + Ag).
Intuitively, this can be understood as a generalization of the
ratio Y/N, which must be >1 for the alignment to be chimeric.
The β parameter (-xn option, which should be >1) gives a no
vote a higher weight than a yes vote, and the n parameter (-dn
option, which should be >0 and is set to 1.4 by default) acts as
a pseudo-count
prior on the number of no votes. A positive value of n reduces
H, especially when Y is small; this models increased uncertainty
with reduced evidence. Abstain votes also lower the score as they
indicate noise or the use of a step-parent, either of which should
increase uncertainty. The query is classified as a chimera if:
H = HL x HR ≥ h.
Here, h is the minimum score threshold (-minh option).
|