See also
Cross-talk
uncross command
This OTU table has 12 samples: three mock, thee human gut, three mouse gut and three soil. The top 25 OTUs generated by UPARSE are shown sorted by decreasing number of reads assigned to the mock samples. Table entries shown in orange are probably due to cross-talk. This is most obvious in five of the last six OTUs which do not match the designed mock community but have high abundance in the gut samples. See below for further discussion.
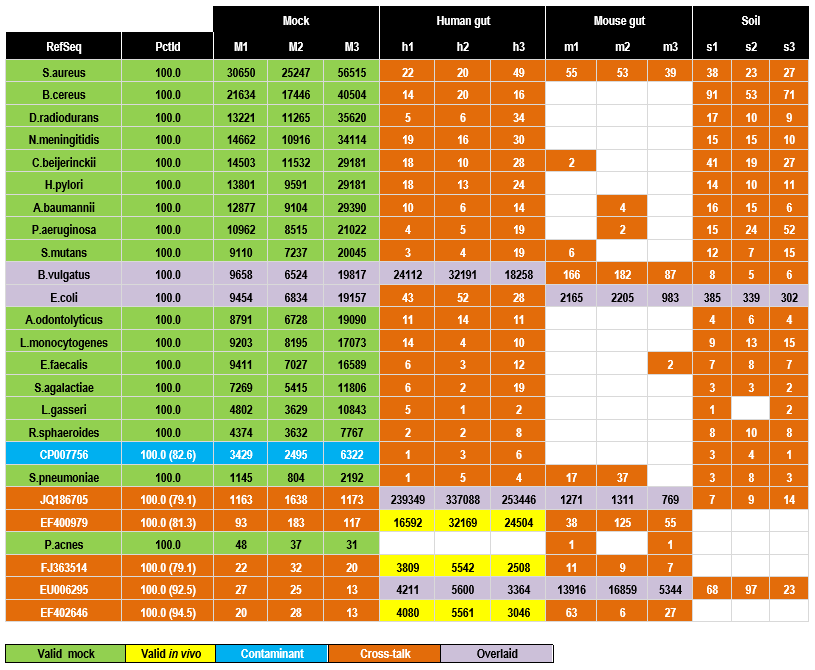
The 25 OTUs from MiSeq run 130417 with highest mock abundances. OTUs are
sorted in order of decreasing total mock abundance. Counts are manually
annotated as valid (green for mock, yellow for environmental), contaminant
(blue, can be detected in mock only), cross-talk (orange) or overlaid
(purple, meaning that the OTU is valid in the mock samples and one or more
environmental sample). Reference sequences with species names (green) are
designed strains in the mock community, otherwise are Genbank identifiers
(blue for contaminant, purple for overlaid and orange for cross-talk). The
PctId column gives identity with the reference sequence, values in
parenthesis are identities with the most similar mock reference sequence to
confirm that the contaminant and cross-talk counts are not due to noisy
reads of expected strains. Note that two cross-talk OTUs (JQ186705 and
EF400979) and one contaminant OTU (CP007756) have higher abundance than the
least abundant expected strain (P. acnes). Data from
Kozich et al. 2013.