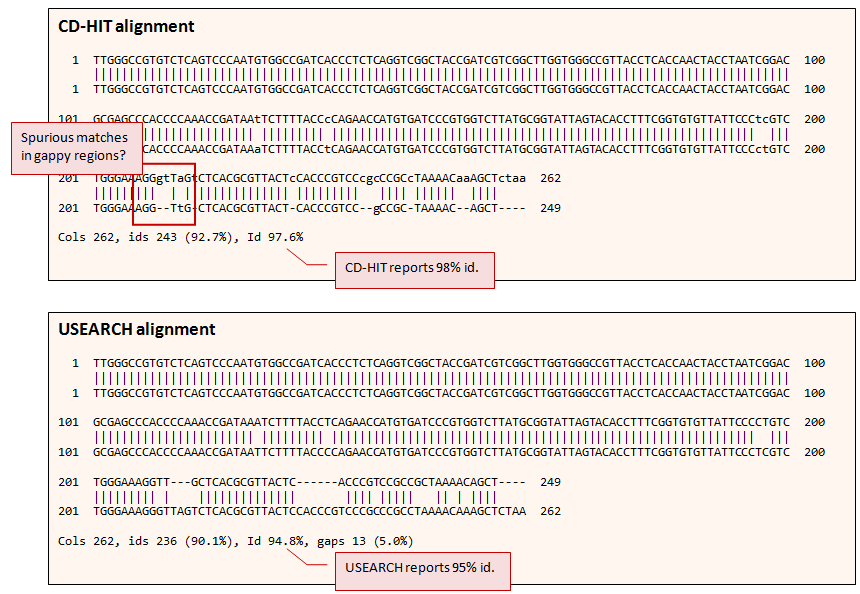
CD-HIT alignments are gappy
<< CD-HIT analysis
<< Comparing USEARCH and CD-HIT
Below are two alignments of a pair of 16S reads (in
FASTA format at bottom of this page). The top alignment is by
CD-HIT-EST v4.5.7, which has 13
gapped columns (not including terminal gaps), and gives an identity of 98% according to the CD-HIT
definition, which does not count gaps in the longer sequence as differences. The lower alignment is by USEARCH, which has 9
internal gapped columns and gives 95% using CD-HIT's measure of identity (--iddef
0 option). See here for instructions on how
to view CD-HIT alignments.
CD-HIT
alignments have spurious matches in gappy regions
Many matches in
gappy regions in CD-HIT alignments are probably
spurious (example in red box below).
Taxonomic
distance
The RDP Naive Bayesian
Classifier assigns both reads to order Clostridiales, with tentative
assignment to the same family (Ruminococcaceae, with P=0.25 for
F12Fcsw_257171 and P=0.48 for M13Fcsw_294419), but different genera.
Since the RDP classifier uses an alignment-free method, we can assume
that it is independent of alignment biases. In this example, the
divergence reported by USEARCH is closer to the expected taxonomic
divergence.
>F12Fcsw_257171
TTGGGCCGTGTCTCAGTCCCAATGTGGCCGATCACCCTCTCAGGTCGGCTACCGATCGTCGGCTTGGTGGGCCGTTACCT
CACCAACTACCTAATCGGACGCGAGCCCACCCCAAACCGATAATTCTTTTACCCCAGAACCATGTGATCCCGTGGTCTTA
TGCGGTATTAGTACACCTTTCGGTGTGTTATTCCCTCGTCTGGGAAAGGGTTAGTCTCACGCGTTACTCCACCCGTCCCG
CCCGCCTAAAACAAAGCTCTAA
>M13Fcsw_294419
TTGGGCCGTGTCTCAGTCCCAATGTGGCCGATCACCCTCTCAGGTCGGCTACCGATCGTCGGCTTGGTGGGCCGTTACCT
CACCAACTACCTAATCGGACGCGAGCCCACCCCAAACCGATAAATCTTTTACCTCAGAACCATGTGATCCCGTGGTCTTA
TGCGGTATTAGTACACCTTTCGGTGTGTTATTCCCCTGTCTGGGAAAGGTTGCTCACGCGTTACTCACCCGTCCGCCGCT
AAAACAGCT
